Dr
Richard Kao Yi-tsun
Associate Professor
Department of Microbiology
My return to Hong Kong
in 2001 to join the Department of Microbiology at The University of Hong Kong
was a great opportunity for me to contribute to the effort to combat SARS and
other emerging and re-emerging infectious diseases. I brought with me expertise
in high-throughput screening (HTS), along with passion for microbiology and
infectious diseases.
With strong
encouragement and support from Professor Yuen Kwok-yung and other colleagues, I
started building an automated HTS platform for screening small molecule
compounds that could become useful tools for studying microbial pathogens and
drugs for combating infectious diseases. This process of massively identifying
chemical compounds for studying living organisms is called "chemical
genetics", then a new concept I had learned and practiced while at Harvard
University. I was fortunate to have come back to Hong Kong and joined HKU just
before the SARS-CoV-1 outbreak so that I could become a member of the department’s
team in combating what was the first pandemic of the 21st century.
In 2003, SARS-CoV-1
caught an unprepared world by total surprise. Borders were shut down and
flights grounded. The world was practically in chaos and economies across the
globe collapsed. Scientists raced to identify the agent causing SARS and
developed measures to combat this newly emerging infection.
The HKU team was
crucial in this battle – the first to identify the causative agent. Right after
the SARS-CoV-1 was identified and sequenced by the team led by Prof Yuen, the
global scientific communities were eager to find a cure for the disease as SARS
patients were dying every day due to the lack of vaccines and specific drugs
for the virus.
In May, 2003, Prof
David Ho, Director of the Aaron Diamond AIDS Research Center (ADARC) in New
York City, came to Hong Kong with the mission to identify and validate the
world’s first fusion peptide drug for blocking the entry of the SARS-CoV-1 to
cells. I was entrusted with the task of working with him in evaluating the
efficacies of various fusion peptides in inhibiting the virus. We worked
side-by-side in a temporarily built Biosafety Level 3 (BSL-3) laboratory at
Queen Mary Hospital to look for potential drugs against SARS infection.
I still vividly
remember how the reporter for the US
Public Broadcasting Service (PBS) FRONTLINE/World program
and her cameraman followed us as we worked together to
find a potential cure for SARS. “What happened today is that after several days
of incubation, this morning we got a chance to look at the cells, and we found
out that some of the peptides actually can inhibit the virus,” I told Simone
the morning when Prof Ho and I found out that some peptides worked beautifully
in inhibiting the virus, “That means that with these peptides, the virus cannot
get into the cell, and so that's extremely exciting for us.”
The story of our
collaboration with Dr Ho continued even after he returned to the US. In 2004,
after closely working with him and his team, Prof Yuen and I published a paper
in Chemistry & Biology that established the world’s first model of
chemical genetics in viruses and illustrated that a chemical genetic approach
could be employed to probe most, if not all, druggable targets of a pathogenic
virus and identify potential drugs at the same time. In that article, our team
identified more than 100 potential drugs targeting the helicase, the protease,
the entry and other unknown targets of SARS-CoV-1 (Figure 1). It was the first
time that massive amounts of potential drugs and drug targets were identified
and validated in a single study.
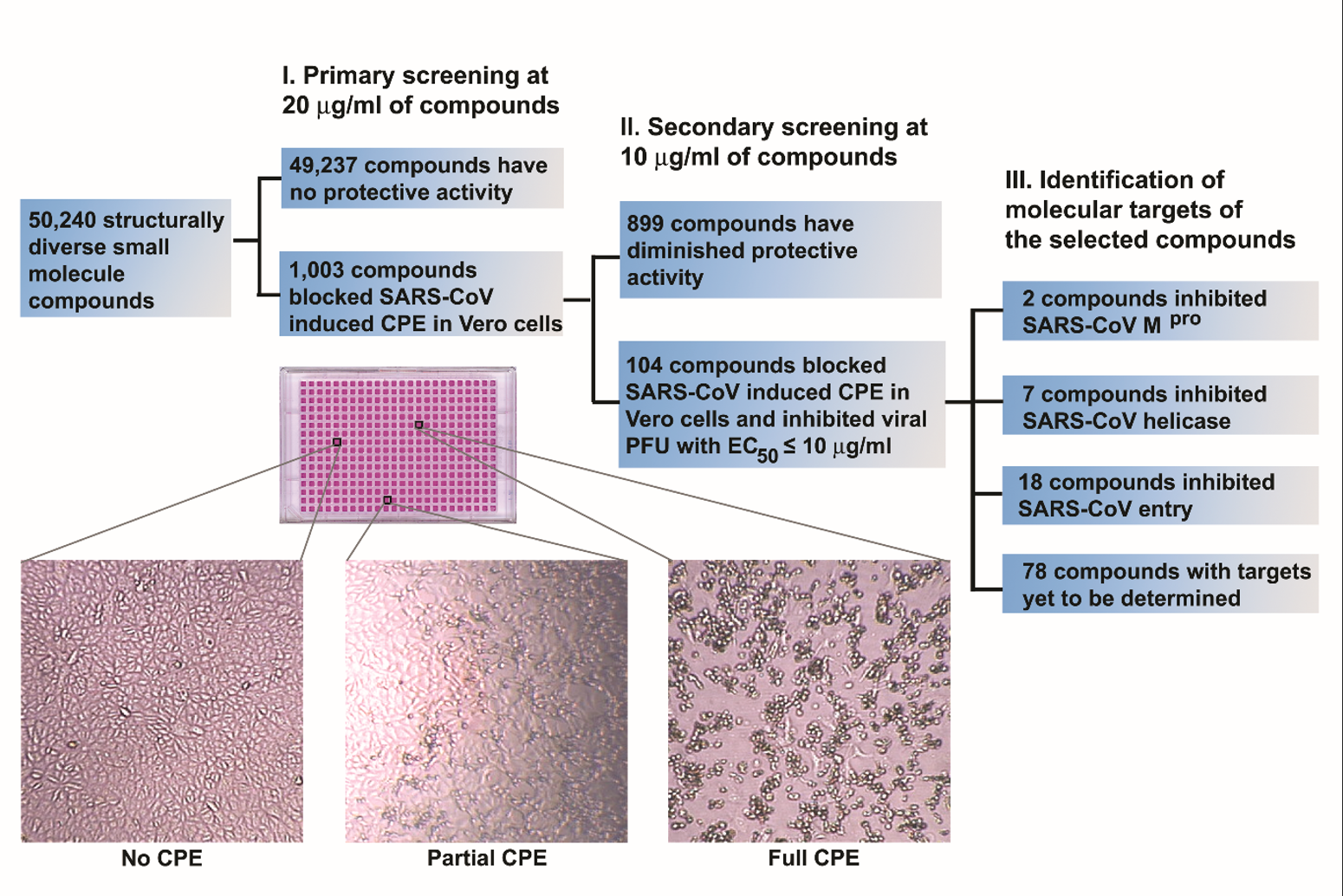
Figure 1. Isolation of biologically active small molecule inhibitors of SARS-CoV-1 in a phenotype-based screen. A schematic illustration of major processes involved in the phenotype-based screen is shown. Enlarged images of Vero cells from a typical 384-well tissue culture plate used in screening are also included to indicate the criteria for hit selection. Only those compounds that fully protected the Vero cells from SARS-CoV induced cytopathic effect (CPE) were selected as hits.
Because of the timely
discovery of many potential drugs for the treatment of SARS-CoV-1 infection, Time magazine
interviewed me for a story
that was published in September 2004: “HKU's work on SARS and bird flu has
helped transform a regional university into a world player in disease research,
and its staff understand that they are part of a vital bulwark.” Two members
from Ho’s laboratory, Chen Zhiwei and Zhang Linqi, were co-authors of the
study. Chen later joined HKU as director of AIDS Institute and now a professor
in the Department of Microbiology. Zhang is a professor at Tsinghua University
in Beijing.
After the SARS-CoV-1
pandemic ended, I employed similar approaches to identify new druggable targets
in influenza viruses and also potential drugs for treating influenza infection.
After screening numerous compounds in cell-based infection assays and
subsequent mechanistic studies, in 2010 I published in Nature Biotechnology an article detailing the ground-breaking discovery of influenza A nucleoprotein
(NP) as a novel druggable antiviral target and a compound which I named
"nucleozin" as a potent antagonist of the nucleoprotein (Figure 2).
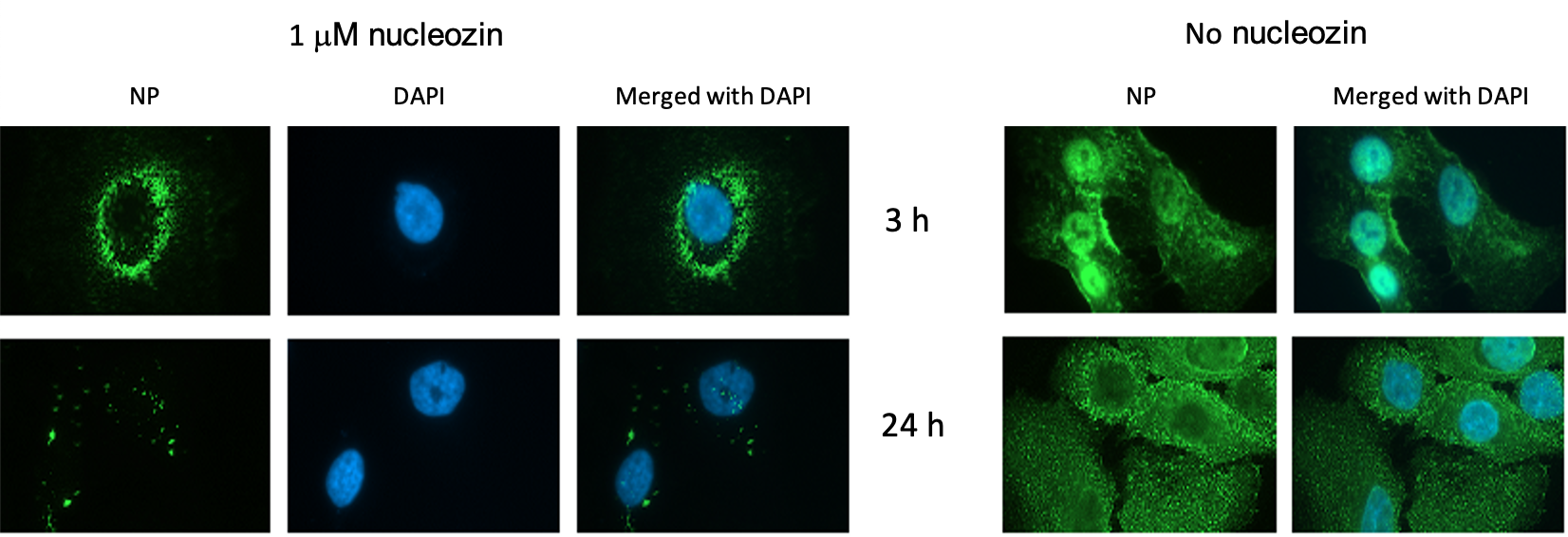
Figure 2. Nucleozin blocked nuclear accumulation of influenza A NP in virus-infected A549 cells. Cells were infected with the A/WSN/33 virus (10 MOI) in the presence or absence of 1μM nucleozin. Influenza A NP accumulated in the nucleus at early infection stage and was distributed exclusively in the cytoplasm at late infection stage in the absence of nucleozin. At the indicated time point, cells were fixed and DAPI staining and mouse anti-influenza A NP antibodies were used to define the locations of the nucleus and viral NP respectively.
This article was the
first Hong Kong-led study published in this prestigious journal. The study on
nucleoprotein as a druggable target has been influential as it opened up new
avenues in exploring viral structural proteins as potential drug targets for
small-molecule chemotherapy. Nucleozin was licensed out to the pharmaceutical
industry for the development of a novel anti-influenza drug.
In the following
decade, using very similar approaches, students, colleagues and I continued to
identify many new inhibitors covering different druggable targets in pandemic
and seasonal influenza viruses including MERS-CoV, Zika virus, EV-71 virus,
Poliovirus, and SARS-CoV-2 virus. I received the HKU Faculty Research Output Prize
in 2010, the Innovation Academy Award from the International Consortium of
Prevention and Control of Infection (ICPIC) in Geneva, Switzerland, in 2017,
and the 2019 State Scientific and Technological Progress Award from the Chinese
government. I am now also engaged in the InnoHK program, working with a team of
researchers led by Prof Yuen at Centre for Virology, Vaccinology and
Therapeutics (CVVT), for the development of effective broad-spectrum antivirals
for the treatment of respiratory viral infections.
created with
WordPress Website Builder .